『医学影像』基于Unet+++实现脊柱MRI定位(上)
发布时间:2025-07-21 编辑:游乐网
本文介绍基于Unet+++实现脊柱MRI定位的项目。因手动选择锥体截面耗时易错,项目将3D数据映射为2D,用深度学习定位L3水平中间轴向切片。处理数据集为PNG格式,定义网络、数据读取类,经训练和验证,测试集平均定位误差为4.0。

基于Unet+++实现脊柱MRI定位-(上)
目前AIstudio已经有许多基于Unet的分割项目,本项目主要介绍分割网络的另外的应用场景,希望对大家的研究有所启发。
0.研究动机
在医学领域,经常需要分析患某种疾病后身体脂肪含量的变化,一般通过选择某个锥体的截面来估计全身的脂肪含量。 常规的方法是通过手动从几百张影像中选择需要的切片(一般为L3),这种方法即耗时又枯燥,稍不注意还容易出错。
在选择到目标切片后,随后进行手动分割,然后使用相关公式估计全身的脂肪含量。
切片选择相关的研究大部分都是在3D数据上对所有的锥体进行标注,但是这个任务中不需要其他的锥体的具体位置,而且3D数据对设备的要求更高。
因此,目前的一个解决方案是通过将三维数据映射使用MLP映射到二维,然后使用深度学习进行定位。
一个经典的解决方案如下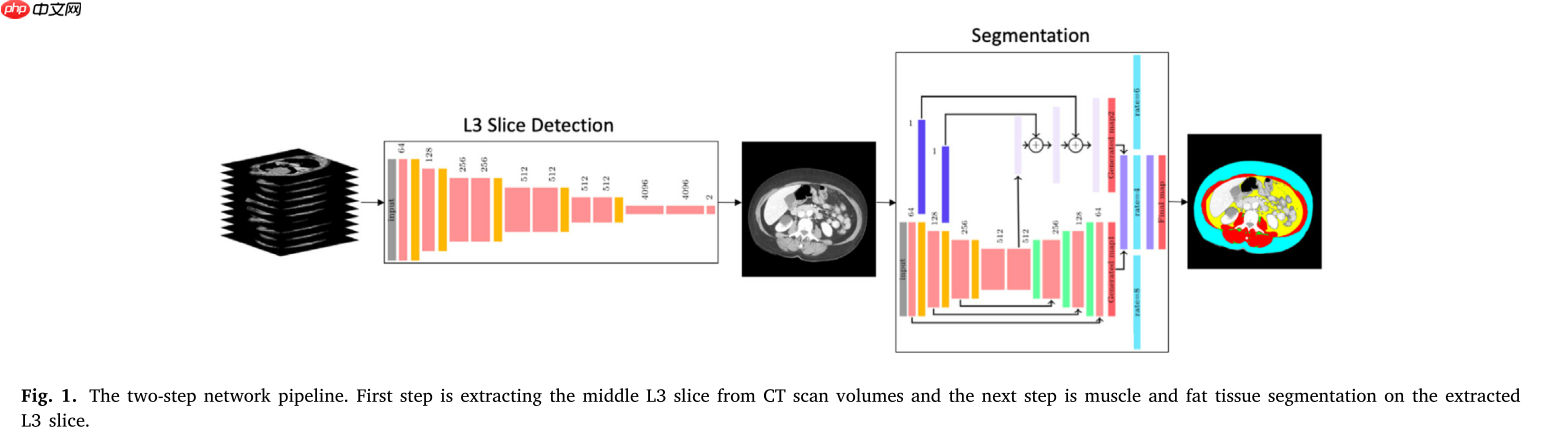
1.项目介绍
计算机断层扫描(CT)成像广泛用于研究身体成分,即肌肉和脂肪组织的比例,应用于营养或化疗剂量设计等领域。
特别是,来自固定位置的轴向CT切片通常用于身体成分分析。然而,如果手动进行,从数百张切片中手动选择是非常繁琐的操作。
本项目的目的是从全身或部分身体扫描体积中自动找到L3水平的中间轴向切片。
2.数据集介绍
使用公开数据集---磁共振图像脊柱结构多类别三维自动分割数据集,该数据集是一个分割数据集,数据格式是nii.gz。分割磁共振T2腰椎矢状位,加背景一共20类。
椎体有S、L5、L4、L3、L2、L1、T12、T11、T10、T9,椎间盘有L5/S, L4/L5, L3/L4, L2/L3, L1/L2, T12/L1, T11/T12, T10/T11, T9/T10
我们对该数据集进行二次处理,包括MLP,剪裁等,建立自己的实验数据集。

3.代码实现
3.1 解压数据并导入常用库
In [ ]# 数据集解压#!unzip -o data/data81211/train.zip -d /home/aistudio/work/登录后复制 In [ ]
#安装 nii处理工具 SimpleITK 和分割工具paddleSeg!pip install SimpleITK!pip install paddleseg!pip install nibabel登录后复制 In [ ]
#导入常用库import osimport randomimport numpy as npimport matplotlib.pyplot as pltfrom random import shuffleimport cv2import paddlefrom PIL import Imageimport shutilimport reimport globimport reimport SimpleITK as sitk登录后复制
3.2 将数据处理为PNG格式
使用分割的思路来解决定位问题,多次实验将目标位置宽度设置为7个像素效果最佳。
slices的选择与窗宽窗位需要自己根据数据调整
In [ ]from PIL import Imagedef read_intensity(path): sitkImage = sitk.ReadImage(path) intensityWindowingFilter = sitk.IntensityWindowingImageFilter() #转换成0到255之间 intensityWindowingFilter.SetOutputMaximum(255) intensityWindowingFilter.SetOutputMinimum(0) if 'mask' not in path: #调窗宽窗位 intensityWindowingFilter.SetWindowMaximum(1900) intensityWindowingFilter.SetWindowMinimum(-300) sitkImage = intensityWindowingFilter.Execute(sitkImage) return sitkImagefilename = r'data//Data_L3Location//'if not os.path.exists(filename): os.mkdir(filename)path_ ='work/train/MR/*.nii.gz'dcm_list_ = glob.glob(path_)s_s = 4 # 开始slices位置s_e = 6 # 结束slices位置idx = 0for i,_ in enumerate(dcm_list_): item = dcm_list_[i] NUM = re.findall("\d+",item)[0] print(i,idx) path_mri ='work/train/MR/Case' + str(NUM) + '.nii.gz' path_mask ='work/train/Mask/mask_case' + str(NUM) + '.nii.gz' mri = read_intensity(path_mri) mask = read_intensity(path_mask) npdata = sitk.GetArrayFromImage(mri) npmask = sitk.GetArrayFromImage(mask) npdata = cv2.flip(np.transpose(npdata[:,:,:],(1,2,0)),0) npmask = cv2.flip(np.transpose(npmask[:,:,:],(1,2,0)),0) h,w = np.max(npdata[:,:,s_s:s_e],2).shape if h<768 or w<696: continue else: scale = 0.3 npdata[:,:int(scale*npdata.shape[1]),:] = 0 npdata[:,int((1-scale)*npdata.shape[1]):,:] = 0 npdata_max = np.max(npdata[:,:,s_s:s_e],2) # 最大值压缩 npdata_mean = np.mean(npdata[:,:,s_s:s_e],2) # 均值压缩 npdata_mix = 0.5*(npdata_max+npdata_mean) # 混合压缩 npmask_ = np.max(npmask[:,:,s_s:s_e],2) npmask_13 = npmask_.copy() npmask_14 = npmask_.copy() # 13 / 14 L3 npmask_13[npmask_ != 13] = 0 npmask_14[npmask_ != 14] = 0 npmask_13[npmask_13 == 13] = 255 npmask_14[npmask_14 == 14] = 255 mid_13 = np.where(np.max(npmask_13,1) == 255)[0].mean() # 获取13的中间行索引 mid_14 = np.where(np.max(npmask_14,1) == 255)[0].mean() # 获取14的中间行索引 mid_index = int((mid_13+mid_14)*0.5) # 获取 L3锥体的中间行索引 # 对数据进行截断 npdata_max = npdata_max[:768,184:696] # 880/2 = 440 - 256 = 184 目的是取中间的512列 npdata_mix = npdata_mix[:768,184:696] # 880/2 = 440 - 256 = 184 目的是取中间的512列 npdata_mean = npdata_mean[:768,184:696] # 880/2 = 440 - 256 = 184 目的是取中间的512列 mask = np.zeros_like(npdata_max) mask[mid_index-3:mid_index+3,int(scale*mask.shape[1]):int((1-scale)*mask.shape[1])] = 255 # 标注 L3锥体的中间位置 # 对数据两侧进行切除处理 img_ma = Image.fromarray(np.uint8(npdata_max)) img_mi = Image.fromarray(np.uint8(npdata_mix)) img_me = Image.fromarray(np.uint8(npdata_mean)) img_la = Image.fromarray(np.uint8(mask)) img_ma.save(filename+'max_'+str(idx) +'.png') img_mi.save(filename+'mix_'+str(idx) +'.png') img_me.save(filename+'mean_'+str(idx) +'.png') img_la.save(filename+'label_'+str(idx) +'.png') idx = idx+1登录后复制 3.3 定义数据读取类
训练集与测试集比例为8:2In [ ]import paddlefrom paddle.io import Datasetimport paddleseg.transforms as Timport matplotlib.image as mpimg # mpimg 用于读取图片import numpy as np# 重写数据读取类class MRILocationDataset(Dataset): def __init__(self,mode = 'train',transform =None): label_path_ ='data/Data_L3Location/label_*.png' self.png_list_ = glob.glob(label_path_) self.transforms = transform self.mode = mode # 选择前80%训练,后20%测试 if self.mode == 'train': self.png_list_ = self.png_list_[:int(0.8*len(self.png_list_))] else: self.png_list_ = self.png_list_[int(0.8*len(self.png_list_)):] def __getitem__(self, index): item = self.png_list_[index] mask = mpimg.imread(item) # 读取和代码处于同一目录下的 lena.png mix_ = mpimg.imread(item.replace('label','mix')) max_ = mpimg.imread(item.replace('label','max')) mean_ = mpimg.imread(item.replace('label','mean')) mask = np.expand_dims(mask, axis=0) mix_ = np.expand_dims(mix_, axis=0) max_ = np.expand_dims(max_, axis=0) mean_ = np.expand_dims(mean_, axis=0) data = np.concatenate((mix_,max_,mean_),axis=0) if self.transforms: data ,mask= self.transforms(data,mask) return data ,mask def __len__(self): return len(self.png_list_)登录后复制 In [ ]# 预览数据dataset = MRILocationDataset(mode='train')print('=============train dataset=============')imga, imgb = dataset[4]print(imga.shape,imgb.shape)imga = imga[0]*255imga = Image.fromarray(imga)#当要保存的图片为灰度图像时,灰度图像的 numpy 尺度是 [1, h, w]。需要将 [1, h, w] 改变为 [h, w]imgb = np.squeeze(imgb)plt.figure(figsize=(12, 6))plt.subplot(1,2,1),plt.xticks([]),plt.yticks([]),plt.imshow(imga)plt.subplot(1,2,2),plt.xticks([]),plt.yticks([]),plt.imshow(imgb)plt.show()登录后复制 =============train dataset=============(3, 768, 512) (1, 768, 512)登录后复制
登录后复制
3.4 定义unet+++网络
简介
UNet的发展
2006年Hinton大神提出了一种encoder-decoder结构,当时这个encoder-decoder结构提出的主要作用并不是分割,而是压缩图像和去噪声。输入是一幅图,经过下采样的编码,得到一串比原先图像更小的特征,相当于压缩,然后再经过一个解码,理想状况就是能还原到原来的图像。而在2015,基于此拓扑结构的FCN和UNet相继提出,其中UNet的对称结构简单易懂,效果还好,就成为了许多网络改进的范本之一。
来源
ICASSP 2020 paper 《UNet 3+: A full-scale connected unet for medical image segmentation》
设计特点
全尺度连接:
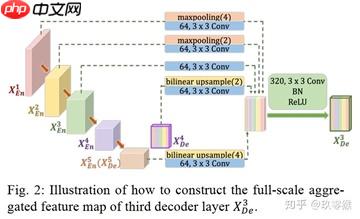
为了弥补UNet和UNet++不能精确分割图像中器官的位置和边界,UNet3+中每一个解码器都结合了全部编码器的特征,这些不同尺度的特征能够获取细粒度的细节和粗粒度的语义。UNet 3+中的每一个解码器层都融合了来自编码器中的小尺度和同尺度的特征图,以及来自解码器的da尺度的特征图,这些特征图捕获了全尺度下的细粒度语义和粗粒度语义。下图表明了第三层解码器的特征图如何构造
全尺度监督:
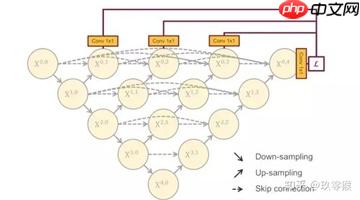
在UNet++中,已经实现了深度监督。它对生成的全分辨率特征图进行操作,即 X0,1 、X0,2、 X0,3 、X0,4后面加一个1x1的卷积核,相当于监督每个分支的UNet的输出。与UNet++对每个嵌套的子网络进行监督不同的是,在UNet3+中每一个解码器模块都有一个输出,与ground truth进行比较计算loss,从而实现全尺度的监督
分类引导模块:
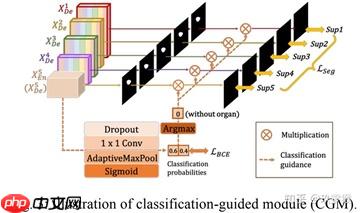
为了防止非器官图像的过度分割,和提高模型的分割精度,作者通过添加一个额外的分类任务来预测输入图像是否有器官,从而实现更精准的分割。具体就是利用最丰富的语义信息,分类结果可以进一步指导每一个切分侧边输出两个步骤。首先,在argmax函数的帮助下,将二维张量转化为{0,1}的单个输出,表示有/没有目标。随后将单个分类输出与侧分割输出相乘。由于二值分类任务的简单性,该模块通过优化二值交叉熵损失函数,轻松获得准确的分类结果,实现了对非目标图像过分割的指导。
(图源知乎:玖零猴,侵删)
网络结构
与UNet和UNet++相比,UNet3+结合了多尺度特征,重新设计了跳跃连接,并利用多尺度的深度监督,UNet3+提供更少的参数,但可以产生更准确的位置感知和边界增强的分割图
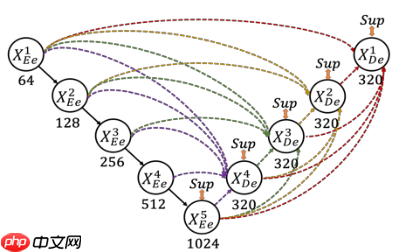
说明
pytorch版本中有UNet3+、用到了深度监督的UNet3+以及分类指导模块的UNet3+,都以在unet.py中转为paddle的版本。具体介绍还是请移步知乎:UNet3+(UNet+++)论文解读
参考项目 https://aistudio.baidu.com/aistudio/projectdetail/1555546In [ ]import paddleimport paddle.nn as nnimport paddle.nn.functional as Ffrom paddle.nn import initializerdef init_weights(init_type='kaiming'): if init_type == 'normal': return paddle.framework.ParamAttr(initializer=paddle.nn.initializer.Normal()) elif init_type == 'xavier': return paddle.framework.ParamAttr(initializer=paddle.nn.initializer.XavierNormal()) elif init_type == 'kaiming': return paddle.framework.ParamAttr(initializer=paddle.nn.initializer.KaimingNormal) else: raise NotImplementedError('initialization method [%s] is not implemented' % init_type)class unetConv2(nn.Layer): def __init__(self, in_size, out_size, is_batchnorm, n=2, ks=3, stride=1, padding=1): super(unetConv2, self).__init__() self.n = n self.ks = ks self.stride = stride self.padding = padding s = stride p = padding if is_batchnorm: for i in range(1, n + 1): conv = nn.Sequential(nn.Conv2D(in_size, out_size, ks, s, p), nn.BatchNorm(out_size), nn.ReLU(), ) setattr(self, 'conv%d' % i, conv) in_size = out_size else: for i in range(1, n + 1): conv = nn.Sequential(nn.Conv2D(in_size, out_size, ks, s, p), nn.ReLU(), ) setattr(self, 'conv%d' % i, conv) in_size = out_size # initialise the blocks for m in self.children(): m.weight_attr=init_weights(init_type='kaiming') m.bias_attr=init_weights(init_type='kaiming') def forward(self, inputs): x = inputs for i in range(1, self.n + 1): conv = getattr(self, 'conv%d' % i) x = conv(x) return x''' UNet 3+'''class UNet_3Plus(nn.Layer): def __init__(self, in_channels=3, n_classes=1, is_deconv=True, is_batchnorm=True, end_sigmoid=True): super(UNet_3Plus, self).__init__() self.is_deconv = is_deconv self.in_channels = in_channels self.is_batchnorm = is_batchnorm self.end_sigmoid = end_sigmoid filters = [16, 32, 64, 128, 256] ## -------------Encoder-------------- self.conv1 = unetConv2(self.in_channels, filters[0], self.is_batchnorm) self.maxpool1 = nn.MaxPool2D(kernel_size=2) self.conv2 = unetConv2(filters[0], filters[1], self.is_batchnorm) self.maxpool2 = nn.MaxPool2D(kernel_size=2) self.conv3 = unetConv2(filters[1], filters[2], self.is_batchnorm) self.maxpool3 = nn.MaxPool2D(kernel_size=2) self.conv4 = unetConv2(filters[2], filters[3], self.is_batchnorm) self.maxpool4 = nn.MaxPool2D(kernel_size=2) self.conv5 = unetConv2(filters[3], filters[4], self.is_batchnorm) ## -------------Decoder-------------- self.CatChannels = filters[0] self.CatBlocks = 5 self.UpChannels = self.CatChannels * self.CatBlocks '''stage 4d''' # h2->320*320, hd4->40*40, Pooling 8 times self.h2_PT_hd4 = nn.MaxPool2D(8, 8, ceil_mode=True) self.h2_PT_hd4_conv = nn.Conv2D(filters[0], self.CatChannels, 3, padding=1) self.h2_PT_hd4_bn = nn.BatchNorm(self.CatChannels) self.h2_PT_hd4_relu = nn.ReLU() # h2->160*160, hd4->40*40, Pooling 4 times self.h2_PT_hd4 = nn.MaxPool2D(4, 4, ceil_mode=True) self.h2_PT_hd4_conv = nn.Conv2D(filters[1], self.CatChannels, 3, padding=1) self.h2_PT_hd4_bn = nn.BatchNorm(self.CatChannels) self.h2_PT_hd4_relu = nn.ReLU() # h3->80*80, hd4->40*40, Pooling 2 times self.h3_PT_hd4 = nn.MaxPool2D(2, 2, ceil_mode=True) self.h3_PT_hd4_conv = nn.Conv2D(filters[2], self.CatChannels, 3, padding=1) self.h3_PT_hd4_bn = nn.BatchNorm(self.CatChannels) self.h3_PT_hd4_relu = nn.ReLU() # h2->40*40, hd4->40*40, Concatenation self.h2_Cat_hd4_conv = nn.Conv2D(filters[3], self.CatChannels, 3, padding=1) self.h2_Cat_hd4_bn = nn.BatchNorm(self.CatChannels) self.h2_Cat_hd4_relu = nn.ReLU() # hd5->20*20, hd4->40*40, Upsample 2 times self.hd5_UT_hd4 = nn.Upsample(scale_factor=2, mode='bilinear') # 14*14 self.hd5_UT_hd4_conv = nn.Conv2D(filters[4], self.CatChannels, 3, padding=1) self.hd5_UT_hd4_bn = nn.BatchNorm(self.CatChannels) self.hd5_UT_hd4_relu = nn.ReLU() # fusion(h2_PT_hd4, h2_PT_hd4, h3_PT_hd4, h2_Cat_hd4, hd5_UT_hd4) self.conv4d_1 = nn.Conv2D(self.UpChannels, self.UpChannels, 3, padding=1) # 16 self.bn4d_1 = nn.BatchNorm(self.UpChannels) self.relu4d_1 = nn.ReLU() '''stage 3d''' # h2->320*320, hd3->80*80, Pooling 4 times self.h2_PT_hd3 = nn.MaxPool2D(4, 4, ceil_mode=True) self.h2_PT_hd3_conv = nn.Conv2D(filters[0], self.CatChannels, 3, padding=1) self.h2_PT_hd3_bn = nn.BatchNorm(self.CatChannels) self.h2_PT_hd3_relu = nn.ReLU() # h2->160*160, hd3->80*80, Pooling 2 times self.h2_PT_hd3 = nn.MaxPool2D(2, 2, ceil_mode=True) self.h2_PT_hd3_conv = nn.Conv2D(filters[1], self.CatChannels, 3, padding=1) self.h2_PT_hd3_bn = nn.BatchNorm(self.CatChannels) self.h2_PT_hd3_relu = nn.ReLU() # h3->80*80, hd3->80*80, Concatenation self.h3_Cat_hd3_conv = nn.Conv2D(filters[2], self.CatChannels, 3, padding=1) self.h3_Cat_hd3_bn = nn.BatchNorm(self.CatChannels) self.h3_Cat_hd3_relu = nn.ReLU() # hd4->40*40, hd4->80*80, Upsample 2 times self.hd4_UT_hd3 = nn.Upsample(scale_factor=2, mode='bilinear') # 14*14 self.hd4_UT_hd3_conv = nn.Conv2D(self.UpChannels, self.CatChannels, 3, padding=1) self.hd4_UT_hd3_bn = nn.BatchNorm(self.CatChannels) self.hd4_UT_hd3_relu = nn.ReLU() # hd5->20*20, hd4->80*80, Upsample 4 times self.hd5_UT_hd3 = nn.Upsample(scale_factor=4, mode='bilinear') # 14*14 self.hd5_UT_hd3_conv = nn.Conv2D(filters[4], self.CatChannels, 3, padding=1) self.hd5_UT_hd3_bn = nn.BatchNorm(self.CatChannels) self.hd5_UT_hd3_relu = nn.ReLU() # fusion(h2_PT_hd3, h2_PT_hd3, h3_Cat_hd3, hd4_UT_hd3, hd5_UT_hd3) self.conv3d_1 = nn.Conv2D(self.UpChannels, self.UpChannels, 3, padding=1) # 16 self.bn3d_1 = nn.BatchNorm(self.UpChannels) self.relu3d_1 = nn.ReLU() '''stage 2d ''' # h2->320*320, hd2->160*160, Pooling 2 times self.h2_PT_hd2 = nn.MaxPool2D(2, 2, ceil_mode=True) self.h2_PT_hd2_conv = nn.Conv2D(filters[0], self.CatChannels, 3, padding=1) self.h2_PT_hd2_bn = nn.BatchNorm(self.CatChannels) self.h2_PT_hd2_relu = nn.ReLU() # h2->160*160, hd2->160*160, Concatenation self.h2_Cat_hd2_conv = nn.Conv2D(filters[1], self.CatChannels, 3, padding=1) self.h2_Cat_hd2_bn = nn.BatchNorm(self.CatChannels) self.h2_Cat_hd2_relu = nn.ReLU() # hd3->80*80, hd2->160*160, Upsample 2 times self.hd3_UT_hd2 = nn.Upsample(scale_factor=2, mode='bilinear') # 14*14 self.hd3_UT_hd2_conv = nn.Conv2D(self.UpChannels, self.CatChannels, 3, padding=1) self.hd3_UT_hd2_bn = nn.BatchNorm(self.CatChannels) self.hd3_UT_hd2_relu = nn.ReLU() # hd4->40*40, hd2->160*160, Upsample 4 times self.hd4_UT_hd2 = nn.Upsample(scale_factor=4, mode='bilinear') # 14*14 self.hd4_UT_hd2_conv = nn.Conv2D(self.UpChannels, self.CatChannels, 3, padding=1) self.hd4_UT_hd2_bn = nn.BatchNorm(self.CatChannels) self.hd4_UT_hd2_relu = nn.ReLU() # hd5->20*20, hd2->160*160, Upsample 8 times self.hd5_UT_hd2 = nn.Upsample(scale_factor=8, mode='bilinear') # 14*14 self.hd5_UT_hd2_conv = nn.Conv2D(filters[4], self.CatChannels, 3, padding=1) self.hd5_UT_hd2_bn = nn.BatchNorm(self.CatChannels) self.hd5_UT_hd2_relu = nn.ReLU() # fusion(h2_PT_hd2, h2_Cat_hd2, hd3_UT_hd2, hd4_UT_hd2, hd5_UT_hd2) self.Conv2D_1 = nn.Conv2D(self.UpChannels, self.UpChannels, 3, padding=1) # 16 self.bn2d_1 = nn.BatchNorm(self.UpChannels) self.relu2d_1 = nn.ReLU() '''stage 1d''' # h2->320*320, hd1->320*320, Concatenation self.h2_Cat_hd1_conv = nn.Conv2D(filters[0], self.CatChannels, 3, padding=1) self.h2_Cat_hd1_bn = nn.BatchNorm(self.CatChannels) self.h2_Cat_hd1_relu = nn.ReLU() # hd2->160*160, hd1->320*320, Upsample 2 times self.hd2_UT_hd1 = nn.Upsample(scale_factor=2, mode='bilinear') # 14*14 self.hd2_UT_hd1_conv = nn.Conv2D(self.UpChannels, self.CatChannels, 3, padding=1) self.hd2_UT_hd1_bn = nn.BatchNorm(self.CatChannels) self.hd2_UT_hd1_relu = nn.ReLU() # hd3->80*80, hd1->320*320, Upsample 4 times self.hd3_UT_hd1 = nn.Upsample(scale_factor=4, mode='bilinear') # 14*14 self.hd3_UT_hd1_conv = nn.Conv2D(self.UpChannels, self.CatChannels, 3, padding=1) self.hd3_UT_hd1_bn = nn.BatchNorm(self.CatChannels) self.hd3_UT_hd1_relu = nn.ReLU() # hd4->40*40, hd1->320*320, Upsample 8 times self.hd4_UT_hd1 = nn.Upsample(scale_factor=8, mode='bilinear') # 14*14 self.hd4_UT_hd1_conv = nn.Conv2D(self.UpChannels, self.CatChannels, 3, padding=1) self.hd4_UT_hd1_bn = nn.BatchNorm(self.CatChannels) self.hd4_UT_hd1_relu = nn.ReLU() # hd5->20*20, hd1->320*320, Upsample 16 times self.hd5_UT_hd1 = nn.Upsample(scale_factor=16, mode='bilinear') # 14*14 self.hd5_UT_hd1_conv = nn.Conv2D(filters[4], self.CatChannels, 3, padding=1) self.hd5_UT_hd1_bn = nn.BatchNorm(self.CatChannels) self.hd5_UT_hd1_relu = nn.ReLU() # fusion(h2_Cat_hd1, hd2_UT_hd1, hd3_UT_hd1, hd4_UT_hd1, hd5_UT_hd1) self.conv1d_1 = nn.Conv2D(self.UpChannels, self.UpChannels, 3, padding=1) # 16 self.bn1d_1 = nn.BatchNorm(self.UpChannels) self.relu1d_1 = nn.ReLU() # output self.outconv1 = nn.Conv2D(self.UpChannels, n_classes, 3, padding=1) # initialise weights for m in self.sublayers (): if isinstance(m, nn.Conv2D): m.weight_attr = init_weights(init_type='kaiming') m.bias_attr = init_weights(init_type='kaiming') elif isinstance(m, nn.BatchNorm): m.param_attr =init_weights(init_type='kaiming') m.bias_attr = init_weights(init_type='kaiming') def forward(self, inputs): ## -------------Encoder------------- h2 = self.conv1(inputs) # h2->320*320*64 h2 = self.maxpool1(h2) h2 = self.conv2(h2) # h2->160*160*128 h3 = self.maxpool2(h2) h3 = self.conv3(h3) # h3->80*80*256 h2 = self.maxpool3(h3) h2 = self.conv4(h2) # h2->40*40*512 h5 = self.maxpool4(h2) hd5 = self.conv5(h5) # h5->20*20*1024 ## -------------Decoder------------- h2_PT_hd4 = self.h2_PT_hd4_relu(self.h2_PT_hd4_bn(self.h2_PT_hd4_conv(self.h2_PT_hd4(h2)))) h2_PT_hd4 = self.h2_PT_hd4_relu(self.h2_PT_hd4_bn(self.h2_PT_hd4_conv(self.h2_PT_hd4(h2)))) h3_PT_hd4 = self.h3_PT_hd4_relu(self.h3_PT_hd4_bn(self.h3_PT_hd4_conv(self.h3_PT_hd4(h3)))) h2_Cat_hd4 = self.h2_Cat_hd4_relu(self.h2_Cat_hd4_bn(self.h2_Cat_hd4_conv(h2))) hd5_UT_hd4 = self.hd5_UT_hd4_relu(self.hd5_UT_hd4_bn(self.hd5_UT_hd4_conv(self.hd5_UT_hd4(hd5)))) hd4 = self.relu4d_1(self.bn4d_1(self.conv4d_1( paddle.concat([h2_PT_hd4, h2_PT_hd4, h3_PT_hd4, h2_Cat_hd4, hd5_UT_hd4], 1)))) # hd4->40*40*UpChannels h2_PT_hd3 = self.h2_PT_hd3_relu(self.h2_PT_hd3_bn(self.h2_PT_hd3_conv(self.h2_PT_hd3(h2)))) h2_PT_hd3 = self.h2_PT_hd3_relu(self.h2_PT_hd3_bn(self.h2_PT_hd3_conv(self.h2_PT_hd3(h2)))) h3_Cat_hd3 = self.h3_Cat_hd3_relu(self.h3_Cat_hd3_bn(self.h3_Cat_hd3_conv(h3))) hd4_UT_hd3 = self.hd4_UT_hd3_relu(self.hd4_UT_hd3_bn(self.hd4_UT_hd3_conv(self.hd4_UT_hd3(hd4)))) hd5_UT_hd3 = self.hd5_UT_hd3_relu(self.hd5_UT_hd3_bn(self.hd5_UT_hd3_conv(self.hd5_UT_hd3(hd5)))) hd3 = self.relu3d_1(self.bn3d_1(self.conv3d_1( paddle.concat([h2_PT_hd3, h2_PT_hd3, h3_Cat_hd3, hd4_UT_hd3, hd5_UT_hd3], 1)))) # hd3->80*80*UpChannels h2_PT_hd2 = self.h2_PT_hd2_relu(self.h2_PT_hd2_bn(self.h2_PT_hd2_conv(self.h2_PT_hd2(h2)))) h2_Cat_hd2 = self.h2_Cat_hd2_relu(self.h2_Cat_hd2_bn(self.h2_Cat_hd2_conv(h2))) hd3_UT_hd2 = self.hd3_UT_hd2_relu(self.hd3_UT_hd2_bn(self.hd3_UT_hd2_conv(self.hd3_UT_hd2(hd3)))) hd4_UT_hd2 = self.hd4_UT_hd2_relu(self.hd4_UT_hd2_bn(self.hd4_UT_hd2_conv(self.hd4_UT_hd2(hd4)))) hd5_UT_hd2 = self.hd5_UT_hd2_relu(self.hd5_UT_hd2_bn(self.hd5_UT_hd2_conv(self.hd5_UT_hd2(hd5)))) hd2 = self.relu2d_1(self.bn2d_1(self.Conv2D_1( paddle.concat([h2_PT_hd2, h2_Cat_hd2, hd3_UT_hd2, hd4_UT_hd2, hd5_UT_hd2], 1)))) # hd2->160*160*UpChannels h2_Cat_hd1 = self.h2_Cat_hd1_relu(self.h2_Cat_hd1_bn(self.h2_Cat_hd1_conv(h2))) hd2_UT_hd1 = self.hd2_UT_hd1_relu(self.hd2_UT_hd1_bn(self.hd2_UT_hd1_conv(self.hd2_UT_hd1(hd2)))) hd3_UT_hd1 = self.hd3_UT_hd1_relu(self.hd3_UT_hd1_bn(self.hd3_UT_hd1_conv(self.hd3_UT_hd1(hd3)))) hd4_UT_hd1 = self.hd4_UT_hd1_relu(self.hd4_UT_hd1_bn(self.hd4_UT_hd1_conv(self.hd4_UT_hd1(hd4)))) hd5_UT_hd1 = self.hd5_UT_hd1_relu(self.hd5_UT_hd1_bn(self.hd5_UT_hd1_conv(self.hd5_UT_hd1(hd5)))) hd1 = self.relu1d_1(self.bn1d_1(self.conv1d_1( paddle.concat([h2_Cat_hd1, hd2_UT_hd1, hd3_UT_hd1, hd4_UT_hd1, hd5_UT_hd1], 1)))) # hd1->320*320*UpChannels d1 = self.outconv1(hd1) # d1->320*320*n_classes if self.end_sigmoid: out = F.sigmoid(d1) else: out = d1 return out登录后复制 In [ ]# 模型可视化import numpyimport paddleunet3p = UNet_3Plus(in_channels=3, n_classes=1)model = paddle.Model(unet3p)model.summary((2,3, 768, 512))登录后复制
--------------------------------------------------------------------------- Layer (type) Input Shape Output Shape Param # =========================================================================== Conv2D-2 [[2, 3, 768, 512]] [2, 16, 768, 512] 448 BatchNorm-1 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-1 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 Conv2D-3 [[2, 16, 768, 512]] [2, 16, 768, 512] 2,320 BatchNorm-2 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-2 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 unetConv2-2 [[2, 3, 768, 512]] [2, 16, 768, 512] 0 MaxPool2D-1 [[2, 16, 768, 512]] [2, 16, 384, 256] 0 Conv2D-4 [[2, 16, 384, 256]] [2, 32, 384, 256] 4,640 BatchNorm-3 [[2, 32, 384, 256]] [2, 32, 384, 256] 128 ReLU-3 [[2, 32, 384, 256]] [2, 32, 384, 256] 0 Conv2D-5 [[2, 32, 384, 256]] [2, 32, 384, 256] 9,248 BatchNorm-4 [[2, 32, 384, 256]] [2, 32, 384, 256] 128 ReLU-4 [[2, 32, 384, 256]] [2, 32, 384, 256] 0 unetConv2-3 [[2, 16, 384, 256]] [2, 32, 384, 256] 0 MaxPool2D-2 [[2, 32, 384, 256]] [2, 32, 192, 128] 0 Conv2D-6 [[2, 32, 192, 128]] [2, 64, 192, 128] 18,496 BatchNorm-5 [[2, 64, 192, 128]] [2, 64, 192, 128] 256 ReLU-5 [[2, 64, 192, 128]] [2, 64, 192, 128] 0 Conv2D-7 [[2, 64, 192, 128]] [2, 64, 192, 128] 36,928 BatchNorm-6 [[2, 64, 192, 128]] [2, 64, 192, 128] 256 ReLU-6 [[2, 64, 192, 128]] [2, 64, 192, 128] 0 unetConv2-4 [[2, 32, 192, 128]] [2, 64, 192, 128] 0 MaxPool2D-3 [[2, 64, 192, 128]] [2, 64, 96, 64] 0 Conv2D-8 [[2, 64, 96, 64]] [2, 128, 96, 64] 73,856 BatchNorm-7 [[2, 128, 96, 64]] [2, 128, 96, 64] 512 ReLU-7 [[2, 128, 96, 64]] [2, 128, 96, 64] 0 Conv2D-9 [[2, 128, 96, 64]] [2, 128, 96, 64] 147,584 BatchNorm-8 [[2, 128, 96, 64]] [2, 128, 96, 64] 512 ReLU-8 [[2, 128, 96, 64]] [2, 128, 96, 64] 0 unetConv2-5 [[2, 64, 96, 64]] [2, 128, 96, 64] 0 MaxPool2D-4 [[2, 128, 96, 64]] [2, 128, 48, 32] 0 Conv2D-10 [[2, 128, 48, 32]] [2, 256, 48, 32] 295,168 BatchNorm-9 [[2, 256, 48, 32]] [2, 256, 48, 32] 1,024 ReLU-9 [[2, 256, 48, 32]] [2, 256, 48, 32] 0 Conv2D-11 [[2, 256, 48, 32]] [2, 256, 48, 32] 590,080 BatchNorm-10 [[2, 256, 48, 32]] [2, 256, 48, 32] 1,024 ReLU-10 [[2, 256, 48, 32]] [2, 256, 48, 32] 0 unetConv2-6 [[2, 128, 48, 32]] [2, 256, 48, 32] 0 MaxPool2D-5 [[2, 16, 768, 512]] [2, 16, 96, 64] 0 Conv2D-12 [[2, 16, 96, 64]] [2, 16, 96, 64] 2,320 BatchNorm-11 [[2, 16, 96, 64]] [2, 16, 96, 64] 64 ReLU-11 [[2, 16, 96, 64]] [2, 16, 96, 64] 0 MaxPool2D-6 [[2, 32, 384, 256]] [2, 32, 96, 64] 0 Conv2D-13 [[2, 32, 96, 64]] [2, 16, 96, 64] 4,624 BatchNorm-12 [[2, 16, 96, 64]] [2, 16, 96, 64] 64 ReLU-12 [[2, 16, 96, 64]] [2, 16, 96, 64] 0 MaxPool2D-7 [[2, 64, 192, 128]] [2, 64, 96, 64] 0 Conv2D-14 [[2, 64, 96, 64]] [2, 16, 96, 64] 9,232 BatchNorm-13 [[2, 16, 96, 64]] [2, 16, 96, 64] 64 ReLU-13 [[2, 16, 96, 64]] [2, 16, 96, 64] 0 Conv2D-15 [[2, 128, 96, 64]] [2, 16, 96, 64] 18,448 BatchNorm-14 [[2, 16, 96, 64]] [2, 16, 96, 64] 64 ReLU-14 [[2, 16, 96, 64]] [2, 16, 96, 64] 0 Upsample-1 [[2, 256, 48, 32]] [2, 256, 96, 64] 0 Conv2D-16 [[2, 256, 96, 64]] [2, 16, 96, 64] 36,880 BatchNorm-15 [[2, 16, 96, 64]] [2, 16, 96, 64] 64 ReLU-15 [[2, 16, 96, 64]] [2, 16, 96, 64] 0 Conv2D-17 [[2, 80, 96, 64]] [2, 80, 96, 64] 57,680 BatchNorm-16 [[2, 80, 96, 64]] [2, 80, 96, 64] 320 ReLU-16 [[2, 80, 96, 64]] [2, 80, 96, 64] 0 MaxPool2D-8 [[2, 16, 768, 512]] [2, 16, 192, 128] 0 Conv2D-18 [[2, 16, 192, 128]] [2, 16, 192, 128] 2,320 BatchNorm-17 [[2, 16, 192, 128]] [2, 16, 192, 128] 64 ReLU-17 [[2, 16, 192, 128]] [2, 16, 192, 128] 0 MaxPool2D-9 [[2, 32, 384, 256]] [2, 32, 192, 128] 0 Conv2D-19 [[2, 32, 192, 128]] [2, 16, 192, 128] 4,624 BatchNorm-18 [[2, 16, 192, 128]] [2, 16, 192, 128] 64 ReLU-18 [[2, 16, 192, 128]] [2, 16, 192, 128] 0 Conv2D-20 [[2, 64, 192, 128]] [2, 16, 192, 128] 9,232 BatchNorm-19 [[2, 16, 192, 128]] [2, 16, 192, 128] 64 ReLU-19 [[2, 16, 192, 128]] [2, 16, 192, 128] 0 Upsample-2 [[2, 80, 96, 64]] [2, 80, 192, 128] 0 Conv2D-21 [[2, 80, 192, 128]] [2, 16, 192, 128] 11,536 BatchNorm-20 [[2, 16, 192, 128]] [2, 16, 192, 128] 64 ReLU-20 [[2, 16, 192, 128]] [2, 16, 192, 128] 0 Upsample-3 [[2, 256, 48, 32]] [2, 256, 192, 128] 0 Conv2D-22 [[2, 256, 192, 128]] [2, 16, 192, 128] 36,880 BatchNorm-21 [[2, 16, 192, 128]] [2, 16, 192, 128] 64 ReLU-21 [[2, 16, 192, 128]] [2, 16, 192, 128] 0 Conv2D-23 [[2, 80, 192, 128]] [2, 80, 192, 128] 57,680 BatchNorm-22 [[2, 80, 192, 128]] [2, 80, 192, 128] 320 ReLU-22 [[2, 80, 192, 128]] [2, 80, 192, 128] 0 MaxPool2D-10 [[2, 16, 768, 512]] [2, 16, 384, 256] 0 Conv2D-24 [[2, 16, 384, 256]] [2, 16, 384, 256] 2,320 BatchNorm-23 [[2, 16, 384, 256]] [2, 16, 384, 256] 64 ReLU-23 [[2, 16, 384, 256]] [2, 16, 384, 256] 0 Conv2D-25 [[2, 32, 384, 256]] [2, 16, 384, 256] 4,624 BatchNorm-24 [[2, 16, 384, 256]] [2, 16, 384, 256] 64 ReLU-24 [[2, 16, 384, 256]] [2, 16, 384, 256] 0 Upsample-4 [[2, 80, 192, 128]] [2, 80, 384, 256] 0 Conv2D-26 [[2, 80, 384, 256]] [2, 16, 384, 256] 11,536 BatchNorm-25 [[2, 16, 384, 256]] [2, 16, 384, 256] 64 ReLU-25 [[2, 16, 384, 256]] [2, 16, 384, 256] 0 Upsample-5 [[2, 80, 96, 64]] [2, 80, 384, 256] 0 Conv2D-27 [[2, 80, 384, 256]] [2, 16, 384, 256] 11,536 BatchNorm-26 [[2, 16, 384, 256]] [2, 16, 384, 256] 64 ReLU-26 [[2, 16, 384, 256]] [2, 16, 384, 256] 0 Upsample-6 [[2, 256, 48, 32]] [2, 256, 384, 256] 0 Conv2D-28 [[2, 256, 384, 256]] [2, 16, 384, 256] 36,880 BatchNorm-27 [[2, 16, 384, 256]] [2, 16, 384, 256] 64 ReLU-27 [[2, 16, 384, 256]] [2, 16, 384, 256] 0 Conv2D-29 [[2, 80, 384, 256]] [2, 80, 384, 256] 57,680 BatchNorm-28 [[2, 80, 384, 256]] [2, 80, 384, 256] 320 ReLU-28 [[2, 80, 384, 256]] [2, 80, 384, 256] 0 Conv2D-30 [[2, 16, 768, 512]] [2, 16, 768, 512] 2,320 BatchNorm-29 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-29 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 Upsample-7 [[2, 80, 384, 256]] [2, 80, 768, 512] 0 Conv2D-31 [[2, 80, 768, 512]] [2, 16, 768, 512] 11,536 BatchNorm-30 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-30 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 Upsample-8 [[2, 80, 192, 128]] [2, 80, 768, 512] 0 Conv2D-32 [[2, 80, 768, 512]] [2, 16, 768, 512] 11,536 BatchNorm-31 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-31 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 Upsample-9 [[2, 80, 96, 64]] [2, 80, 768, 512] 0 Conv2D-33 [[2, 80, 768, 512]] [2, 16, 768, 512] 11,536 BatchNorm-32 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-32 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 Upsample-10 [[2, 256, 48, 32]] [2, 256, 768, 512] 0 Conv2D-34 [[2, 256, 768, 512]] [2, 16, 768, 512] 36,880 BatchNorm-33 [[2, 16, 768, 512]] [2, 16, 768, 512] 64 ReLU-33 [[2, 16, 768, 512]] [2, 16, 768, 512] 0 Conv2D-35 [[2, 80, 768, 512]] [2, 80, 768, 512] 57,680 BatchNorm-34 [[2, 80, 768, 512]] [2, 80, 768, 512] 320 ReLU-34 [[2, 80, 768, 512]] [2, 80, 768, 512] 0 Conv2D-36 [[2, 80, 768, 512]] [2, 1, 768, 512] 721 ===========================================================================Total params: 1,693,537Trainable params: 1,687,009Non-trainable params: 6,528---------------------------------------------------------------------------Input size (MB): 9.00Forward/backward pass size (MB): 8980.50Params size (MB): 6.46Estimated Total Size (MB): 8995.96---------------------------------------------------------------------------登录后复制
{'total_params': 1693537, 'trainable_params': 1687009}登录后复制 3.5 开始训练
In [14]model = UNet_3Plus(in_channels=3, n_classes=1)#SEUNet(3,1)# 开启模型训练模式model.train()# 定义优化算法,使用随机梯度下降SGD,学习率设置为0.01scheduler = paddle.optimizer.lr.StepDecay(learning_rate=0.01, step_size=30, gamma=0.1, verbose=False)optimizer = paddle.optimizer.Adam(learning_rate=scheduler, parameters=model.parameters())EPOCH_NUM = 60 # 设置外层循环次数BATCH_SIZE = 2 # 设置batch大小train_dataset = MRILocationDataset(mode='train')test_dataset = MRILocationDataset(mode='test')# 使用paddle.io.DataLoader 定义DataLoader对象用于加载Python生成器产生的数据,data_loader = paddle.io.DataLoader(train_dataset, batch_size=BATCH_SIZE, shuffle=False)test_data_loader = paddle.io.DataLoader(test_dataset, batch_size=BATCH_SIZE, shuffle=False)loss_BCEloss = paddle.nn.BCELoss()# 定义外层循环for epoch_id in range(EPOCH_NUM): # 定义内层循环 for iter_id, data in enumerate(data_loader()): x, y = data # x 为数据 ,y 为标签 # 将numpy数据转为飞桨动态图tensor形式 x = paddle.to_tensor(x,dtype='float32') y = paddle.to_tensor(y,dtype='float32') # 前向计算 predicts = model(x) # 计算损失 loss = loss_BCEloss(predicts, y) # 清除梯度 optimizer.clear_grad() # 反向传播 loss.backward() # 最小化loss,更新参数 optimizer.step() scheduler.step() print("epoch: {}, iter: {}, loss is: {}".format(epoch_id+1, iter_id+1, loss.numpy()))# 保存模型参数,文件名为Unet_model.pdparamspaddle.save(model.state_dict(), 'work/Unet3p_model.pdparams')print("模型保存成功,模型参数保存在Unet3p_model.pdparams中")登录后复制 3.6 模型验证
In [15]import paddle# 模型验证Error = []# 清理缓存print("开始测试")# 用于加载之前的训练过的模型参数para_state_dict = paddle.load('work/Unet3p_model.pdparams')model = UNet_3Plus(in_channels=3, n_classes=1)#SEUNet(3,1)model.set_dict(para_state_dict)for iter_id, data in enumerate(test_data_loader()): x, y = data # 将numpy数据转为飞桨动态图tensor形式 x = paddle.to_tensor(x) y = paddle.to_tensor(y) predicts = model(x) for i in range(predicts.shape[0]): predict = predicts[i,:,:,:].cpu().numpy() label = y[i,:,:,:].cpu().numpy() inputs = x[i,1,:,:].cpu().numpy() predict = np.squeeze(predict) label = np.squeeze(label) inputs = np.squeeze(inputs) #当要保存的图片为灰度图像时,灰度图像的 numpy 尺度是 [1, h, w]。需要将 [1, h, w] 改变为 [h, w] plt.figure(figsize=(18, 6)) plt.subplot(1,3,1),plt.xticks([]),plt.yticks([]),plt.imshow(predict) plt.subplot(1,3,2),plt.xticks([]),plt.yticks([]),plt.imshow(label) plt.subplot(1,3,3),plt.xticks([]),plt.yticks([]),plt.imshow(inputs) plt.show() index_predict= np.argmax(np.max(predict,1))+3 index_label = np.argmax(np.max(label,1)) print('真实位置:',index_label,'预测位置:',index_predict) Error.append(np.abs(index_label-index_predict)) breakprint("模型测试集平均定位误差为:",np.mean(Error))登录后复制 开始测试登录后复制
登录后复制登录后复制
真实位置: 416 预测位置: 420登录后复制
登录后复制登录后复制
真实位置: 484 预测位置: 480模型测试集平均定位误差为: 4.0登录后复制
相关阅读
MORE
+- Monodepth2-基于自监督学习的单目深度估计模型 07-22 怎样用豆包AI进行推荐系统开发?协同过滤实战 07-22
- 2021 CCF BDCI基于飞桨实现花样滑冰选手骨骼点动作识别-B榜第3名方案 07-22 FRN——小样本学习SOTA模型 07-22
- deepseek界面操作说明 deepseek怎么调出专业设置 07-22 AI Overviews怎么下载不了 AI Overviews安装失败的常见解决方案 07-22
- Gemini AI怎么用来写简历 Gemini AI自动生成求职内容的方法 07-22 豆包 AI能不能导入本地文件 豆包 AI读取外部文档的操作步骤 07-22
- DeepSeek AI能不能多窗口操作 DeepSeek AI同时处理多个任务的方法 07-22 DeepSeek AI怎么下载离线版 DeepSeek AI本地使用方式是否可行 07-22
- ftp扫描工具排行榜 ftp扫描工具用户评价 07-22 deepseek如何精准搜索 deepseek使用中常见问题解答 07-22
- 基于飞桨复现RealESRGAN 07-22 【方案分享】第十一届 “中国软件杯”大学生软件设计大赛遥感解译赛道 比赛方案分享 07-22
- 豆包AI编程入门指南 豆包AI代码生成方法 07-22 豆包AI+DeepSeek组合潜力挖掘:10个意想不到的实用场景分享 07-22
- 如何用夸克搜索批量查找网课资源 夸克搜索在线学习平台筛选方法 07-22 如何用夸克搜索锁定PDF格式文档 夸克搜索文件类型过滤技巧 07-22





























 湘公网安备
43070202000716号
湘公网安备
43070202000716号